Note
This page was generated from scgen_batch_removal.ipynb. Interactive online version: .
SCGEN: Batch-Removal¶
[1]:
import sys
#if branch is stable, will install via pypi, else will install from source
branch = "stable"
IN_COLAB = "google.colab" in sys.modules
if IN_COLAB and branch == "stable":
!pip install --quiet scgen[tutorials]
elif IN_COLAB and branch != "stable":
!pip install --quiet --upgrade jsonschema
!pip install --quiet git+https://github.com/theislab/scgen@$branch#egg=scgen[tutorials]
|████████████████████████████████| 72 kB 1.0 MB/s
ERROR: pip's dependency resolver does not currently take into account all the packages that are installed. This behaviour is the source of the following dependency conflicts.
nbclient 0.5.12 requires jupyter-client>=6.1.5, but you have jupyter-client 5.3.5 which is incompatible.
Installing build dependencies ... done
Getting requirements to build wheel ... done
Preparing wheel metadata ... done
|████████████████████████████████| 2.0 MB 9.1 MB/s
|████████████████████████████████| 96 kB 6.0 MB/s
|████████████████████████████████| 260 kB 51.3 MB/s
|████████████████████████████████| 8.8 MB 27.7 MB/s
|████████████████████████████████| 4.8 MB 48.3 MB/s
|████████████████████████████████| 1.4 MB 40.0 MB/s
|████████████████████████████████| 48 kB 6.0 MB/s
|████████████████████████████████| 86 kB 5.8 MB/s
|████████████████████████████████| 224 kB 57.8 MB/s
|████████████████████████████████| 283 kB 62.0 MB/s
|████████████████████████████████| 397 kB 59.1 MB/s
|████████████████████████████████| 527 kB 60.3 MB/s
|████████████████████████████████| 713 kB 63.2 MB/s
|████████████████████████████████| 136 kB 62.1 MB/s
|████████████████████████████████| 176 kB 63.7 MB/s
|████████████████████████████████| 829 kB 63.7 MB/s
|████████████████████████████████| 596 kB 53.1 MB/s
|████████████████████████████████| 952 kB 48.3 MB/s
|████████████████████████████████| 134 kB 65.6 MB/s
|████████████████████████████████| 1.1 MB 38.6 MB/s
|████████████████████████████████| 51 kB 7.2 MB/s
|████████████████████████████████| 97 kB 7.2 MB/s
|████████████████████████████████| 1.1 MB 54.7 MB/s
|████████████████████████████████| 144 kB 56.3 MB/s
|████████████████████████████████| 94 kB 3.5 MB/s
|████████████████████████████████| 271 kB 59.4 MB/s
|████████████████████████████████| 3.1 MB 41.2 MB/s
|████████████████████████████████| 70 kB 9.7 MB/s
|████████████████████████████████| 63 kB 1.6 MB/s
Building wheel for scgen (PEP 517) ... done
Building wheel for loompy (setup.py) ... done
Building wheel for docrep (setup.py) ... done
Building wheel for future (setup.py) ... done
Building wheel for umap-learn (setup.py) ... done
Building wheel for pynndescent (setup.py) ... done
Building wheel for adjustText (setup.py) ... done
Building wheel for numpy-groupies (setup.py) ... done
Building wheel for sinfo (setup.py) ... done
ERROR: pip's dependency resolver does not currently take into account all the packages that are installed. This behaviour is the source of the following dependency conflicts.
tensorflow 2.8.0 requires tf-estimator-nightly==2.8.0.dev2021122109, which is not installed.
datascience 0.10.6 requires folium==0.2.1, but you have folium 0.8.3 which is incompatible.
[2]:
import scanpy as sc
import scgen
Global seed set to 0
Loading Train Data¶
[3]:
train = sc.read(
"./tests/data/pancreas.h5ad",
backup_url="https://www.dropbox.com/s/qj1jlm9w10wmt0u/pancreas.h5ad?dl=1"
)
/usr/local/lib/python3.7/dist-packages/anndata/compat/__init__.py:235: FutureWarning: Moving element from .uns['neighbors']['distances'] to .obsp['distances'].
This is where adjacency matrices should go now.
FutureWarning,
/usr/local/lib/python3.7/dist-packages/anndata/compat/__init__.py:235: FutureWarning: Moving element from .uns['neighbors']['connectivities'] to .obsp['connectivities'].
This is where adjacency matrices should go now.
FutureWarning,
We need two observation labels “batch” and “cell_type” for our batch_removal procedure. There exist a “batch” obs but no “cell_type”, so we add it as a .obs of adata
[5]:
train.obs["cell_type"] = train.obs["celltype"].tolist()
sc.pp.neighbors(train)
sc.tl.umap(train)
/usr/local/lib/python3.7/dist-packages/numba/np/ufunc/parallel.py:363: NumbaWarning: The TBB threading layer requires TBB version 2019.5 or later i.e., TBB_INTERFACE_VERSION >= 11005. Found TBB_INTERFACE_VERSION = 9107. The TBB threading layer is disabled.
warnings.warn(problem)
[6]:
sc.pl.umap(train, color=["batch", "cell_type"], wspace=.5, frameon=False)
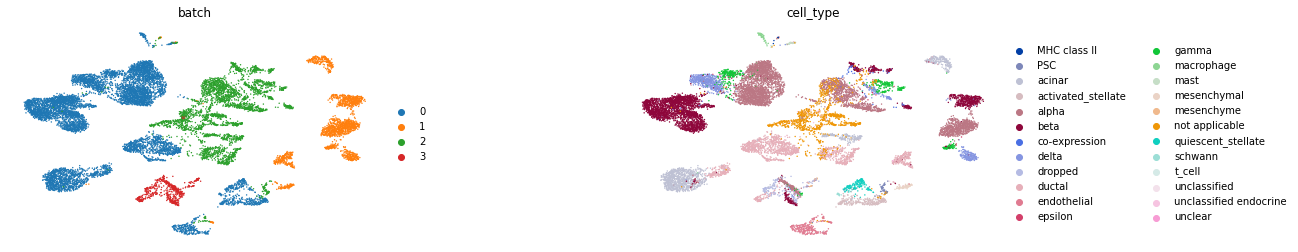
Preprocessing Data¶
[7]:
scgen.SCGEN.setup_anndata(train, batch_key="batch", labels_key="cell_type")
/usr/local/lib/python3.7/dist-packages/scvi/data/_utils.py:158: UserWarning: Category 21 in adata.obs['_scvi_labels'] has fewer than 3 cells. Models may not train properly.
category, alternate_column_key
Creating and Saving the model¶¶
[8]:
model = scgen.SCGEN(train)
model.save("../saved_models/model_batch_removal.pt", overwrite=True)
Training the Model¶
[9]:
model.train(
max_epochs=100,
batch_size=32,
early_stopping=True,
early_stopping_patience=25,
)
GPU available: True, used: True
TPU available: False, using: 0 TPU cores
IPU available: False, using: 0 IPUs
LOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0]
Epoch 27/100: 27%|██▋ | 27/100 [02:36<07:01, 5.78s/it, loss=774, v_num=1]
Monitored metric elbo_validation did not improve in the last 25 records. Best score: 2336.101. Signaling Trainer to stop.
Batch-Removal¶
[10]:
corrected_adata = model.batch_removal()
corrected_adata
/usr/local/lib/python3.7/dist-packages/anndata/_core/anndata.py:1785: FutureWarning: X.dtype being converted to np.float32 from float64. In the next version of anndata (0.9) conversion will not be automatic. Pass dtype explicitly to avoid this warning. Pass `AnnData(X, dtype=X.dtype, ...)` to get the future behavour.
[AnnData(sparse.csr_matrix(a.shape), obs=a.obs) for a in all_adatas],
INFO Input AnnData not setup with scvi-tools. attempting to transfer AnnData setup
/usr/local/lib/python3.7/dist-packages/scvi/data/_utils.py:158: UserWarning: Category 21 in adata.obs['_scvi_labels'] has fewer than 3 cells. Models may not train properly.
category, alternate_column_key
[10]:
AnnData object with n_obs × n_vars = 14693 × 2448
obs: 'celltype', 'sample', 'n_genes', 'batch', 'n_counts', 'louvain', 'cell_type', '_scvi_batch', '_scvi_labels', 'concat_batch'
uns: '_scvi_uuid', '_scvi_manager_uuid'
obsm: 'latent', 'corrected_latent'
Visualization of the corrected gene expression data¶¶
[11]:
sc.pp.neighbors(corrected_adata)
sc.tl.umap(corrected_adata)
sc.pl.umap(corrected_adata, color=['batch', 'cell_type'], wspace=0.4, frameon=False)
WARNING: You’re trying to run this on 2448 dimensions of `.X`, if you really want this, set `use_rep='X'`.
Falling back to preprocessing with `sc.pp.pca` and default params.
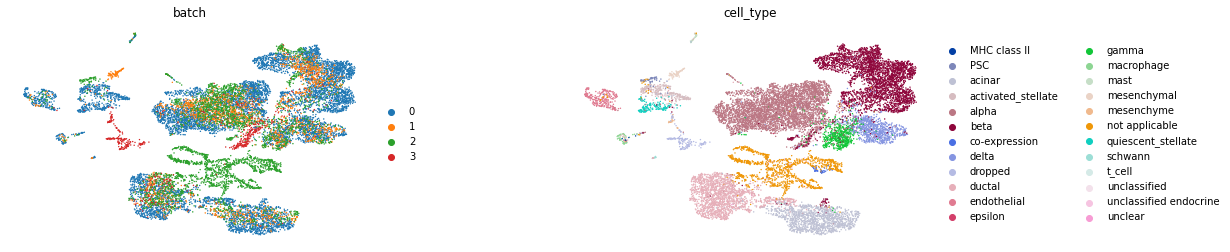
We can also use low-dim corrected gene expression data
[12]:
sc.pp.neighbors(corrected_adata, use_rep="corrected_latent")
sc.tl.umap(corrected_adata)
sc.pl.umap(corrected_adata, color=['batch', 'cell_type'], wspace=0.4, frameon=False)
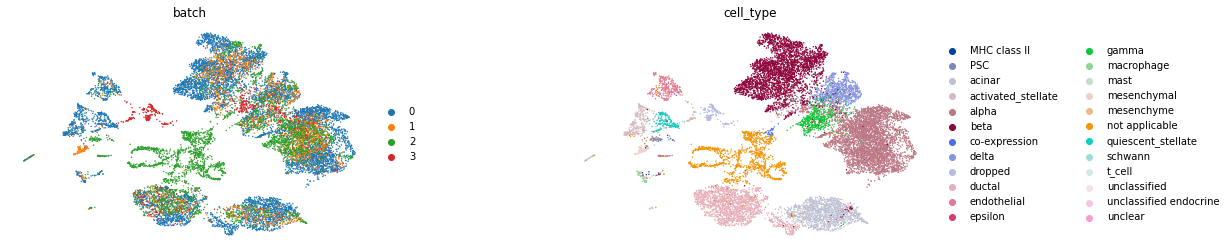
Using Uncorrected Data¶
Note that original adata.raw for the adata.raw is saved to corrected_adata.raw and you can use that for fruther analaysis
[13]:
corrected_adata.raw
[13]:
<anndata._core.raw.Raw at 0x7f4bfc88c7d0>
[14]:
sc.pl.umap(corrected_adata, color=["INS", "cell_type"], wspace=.5, frameon=False, use_raw=True)
